Our Future Bucket List includes a “Personal DNA analyzer.” But once such a thing exists, what will people be able to do with it?
We’re starting on a project that we hope may both answer that question and also help to make such a thing a reality, and part of a much more evenly distributed Future, someday.
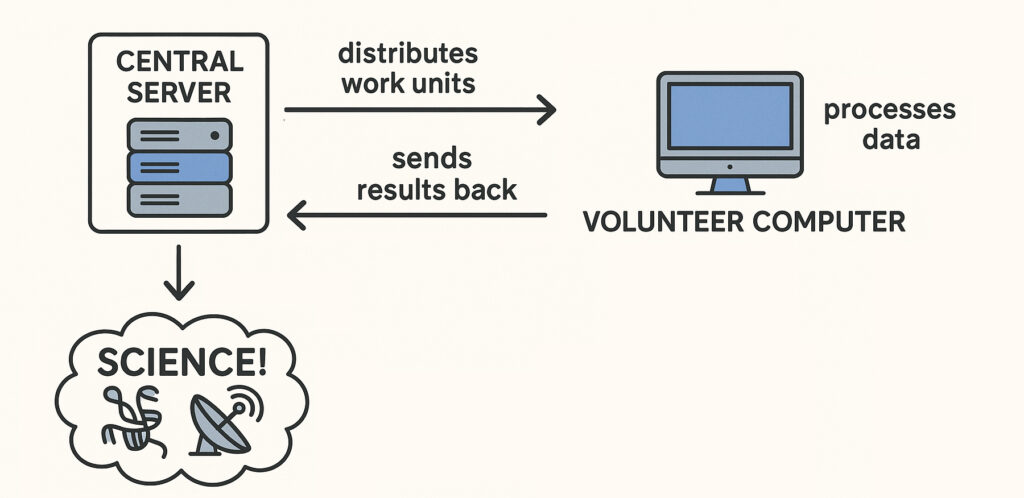
The internet has been a boon to science in many ways. One in particular is “crowdsourcing.” SETI@home utilized volunteers’ internet-connected home computers in the Search for Extra-Terrestrial Intelligence (SETI). Folding@home similarly worked on the difficult but important problem of figuring out how a protein will fold based on the DNA in the gene that codes for that protein.
Sequencing@home would be a great answer to our question about what to do with a personal DNA analyzer. In fact it might even be a reason to create one in the first place.
What would Sequencing@home do? Extrapolating from SETI@home and Folding@home, Sequencing@home would utilize internet-connected home computers to determine (map) the DNA sequence (genome) of the world’s millions of species.
A draft of the first human genome was published, after 10 years and $3 billion, in 2001. Since that time millions of us have had our genomes mapped through projects like the Personal Genome Project. And scientists have sequenced and published the genomes of thousands of major species from around the world. Moore’s law has finally made such projects practical, 30+ years after the Human Genome Project first started.
As amazing as these achievements are, they are just a drop in the proverbial bucket list of sequencing projects. To date, scientists have catalogued and classified millions of earth’s species. In the Future, each species’ entry in this “book of life” could ultimately include the species’ full genome.
That’s where Sequencing@home can come in. As with other crowdsourced efforts, a Sequencing@home central organization would coordinate the effort, distributing the DNA of species to be sequenced to individuals who volunteer their computers, and their time, to the project. The organization would then collect and catalog the results.
There is one big challenge with Sequencing@home when compared to other science @home projects: what’s distributed to the volunteer computers in Sequencing@home would be physical DNA, not electronic data. So it would have to be sent physically, not over the internet (shades of Netflix DVDs?)
This difference also brings us back to the question we started with: the volunteers would need personal DNA analyzers. Most crowdsourcing efforts only require their participants to have an internet-connected computer and a cursory knowledge of the subject at hand. The central organization does the rest. However in this case, what’s being crowdsourced is the actual sequencing, converting a species’ DNA to the (often) billions of letters in that species’ genome.
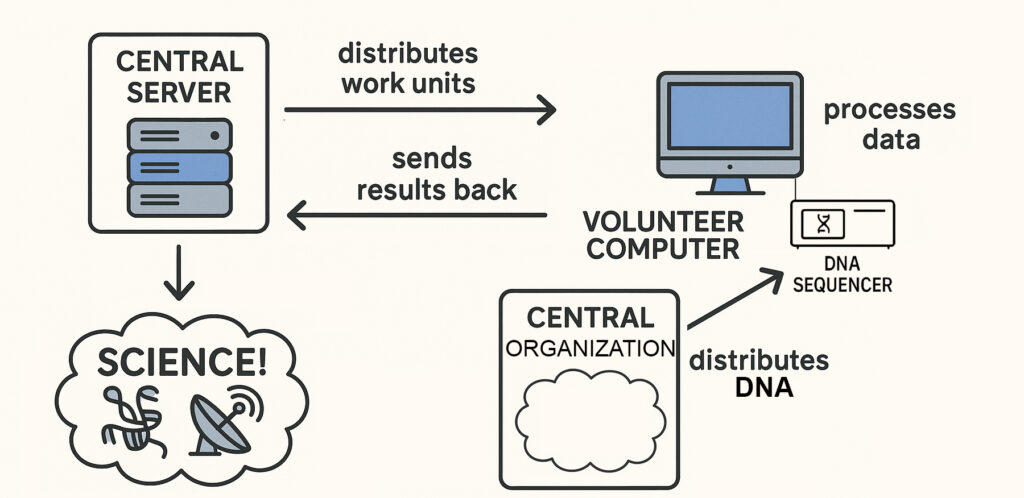
Until very recently, DNA sequencing has been done by commercial DNA analyzers (sequencers), which are large expensive machines run by expert technicians. Very similar to what computers were until the 1970s. Just as computers became personal then, DNA sequencers are just starting to become personal now. And just as key applications on early personal computers (think VisiCalc) drove adoption of those now-essential devices, so too might Sequencing@home drive adoption of personal DNA analyzers. Someday.
As mentioned previously, the Alan & Priscilla Oppenheimer Foundation purchased a $1000 DNA sequencer and is working with Southern Oregon University on a number of genomics-related projects. Not Sequencing@home yet by any means, but perhaps the first step in the 1000 mile journey that might get us to that distributed Future.